在线查询:https://www.jingege.wang/bioinformatics/gene_alias.html
首先肯定是需要自行搜索了解 entrez gene ID, HUGO symbol, refseq ID, ensembl ID 这些专有名词。
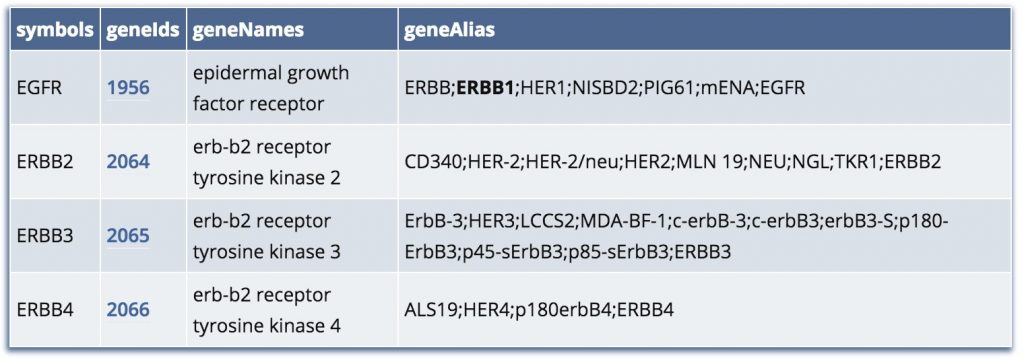
然后直接把下面的代码运行一下,把输出的all_gene_bioconductor.html文件好好看看, 就明白了。
rm(list=ls())
library(org.Hs.eg.db)
eg2symbol=toTable(org.Hs.egSYMBOL)
eg2name=toTable(org.Hs.egGENENAME)
eg2alias=toTable(org.Hs.egALIAS2EG)
eg2alis_list=lapply(split(eg2alias,eg2alias$gene_id),function(x){paste0(x[,2],collapse = ";")})
GeneList=mappedLkeys(org.Hs.egSYMBOL)
if( GeneList[1] %in% eg2symbol$symbol ){
symbols=GeneList
geneIds=eg2symbol[match(symbols,eg2symbol$symbol),'gene_id']
}else{
geneIds=GeneList
symbols=eg2symbol[match(geneIds,eg2symbol$gene_id),'symbol']
}
geneNames=eg2name[match(geneIds,eg2name$gene_id),'gene_name']
geneAlias=sapply(geneIds,function(x){ifelse(is.null(eg2alis_list[[x]]),"no_alias",eg2alis_list[[x]])})
createLink <- function(base,val) {
sprintf('<a href="%s" class="btn btn-link" target="_blank" >%s</a>',base,val)
## target="_blank"
}
gene_info=data.frame( symbols=symbols,
geneIds=createLink(paste0("http://www.ncbi.nlm.nih.gov/gene/",geneIds),geneIds),
geneNames=geneNames,
geneAlias=geneAlias,
stringsAsFactors = F
)
#library("xtable")
#print(xtable(gene_info), type="html",include.rownames=F, file='all_gene.anno',sanitize.text.function = force)
file='all_gene_bioconductor.html'
y <- DT::datatable(gene_info,escape = F,rownames=F)
DT::saveWidget(y,file)
在输出的文件里面可以搜索: