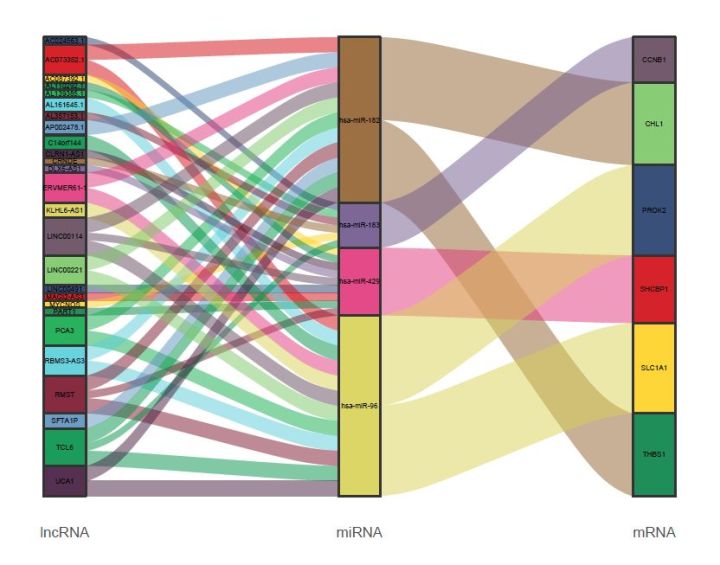
桑基图主要用来展示数据的“流动”变化,分支的宽度表示流量的大小,应用于能源流向、收入支出、人员流动,生物群落丰度变化等数据的可视化。但在之前提到的一篇文献中,作者巧妙地用桑基图来可视化差异ceRNA共表达网络,非常直观。作者构建的 ceRNA network包括26 个lncRNA, 4个miRNAs, 6个 mRNAs,如下:
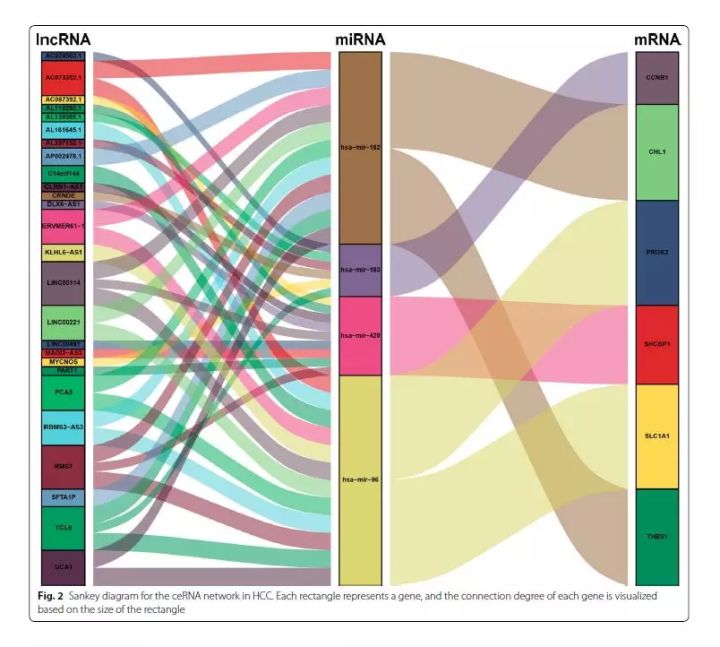 Cancer Cell International, 2019
Cancer Cell International, 2019
文章作者用到的R包是ggalluvial,安装方法非常简单,直接用install.packages(“ggalluvial”)函数在线安装即可。我这里就用这个R包为大家重现原文中的桑基图吧!
数据准备
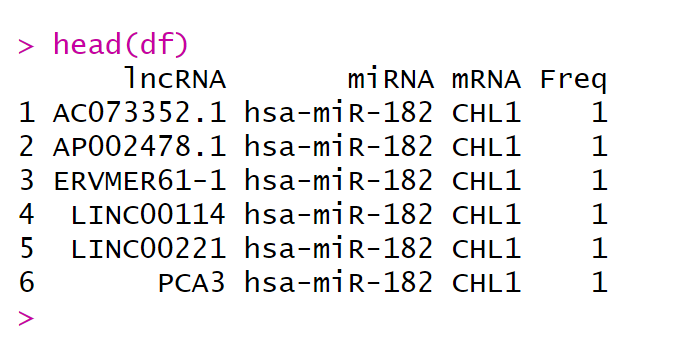
本文用到的范例数据来自原文的附录文件(Table S4),
下载链接:https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6458652/
#载入ggalluvial包;
library(ggalluvial)
#读入数据;
df <- read.table("network.txt",sep= "\t",header = T)
#检查数据的前6行;
head(df)
“宽数据”作图
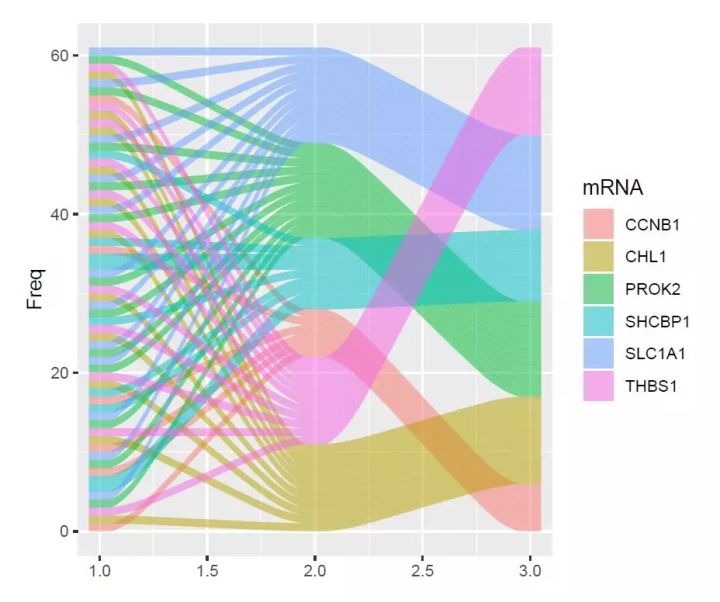
# 直接使用原始数据(宽数据)绘图;
# 绘制alluvium,width调整结点处条带水平宽度;knot.pos调整曲率;reverse调整着色顺序;
p1<-ggplot(data = df,aes(axis1 = lncRNA,axis2 = miRNA,axis3 = mRNA, y = Freq))+
geom_alluvium(aes(fill = mRNA),width = 0.1, knot.pos = 0.1, reverse = F)
p1
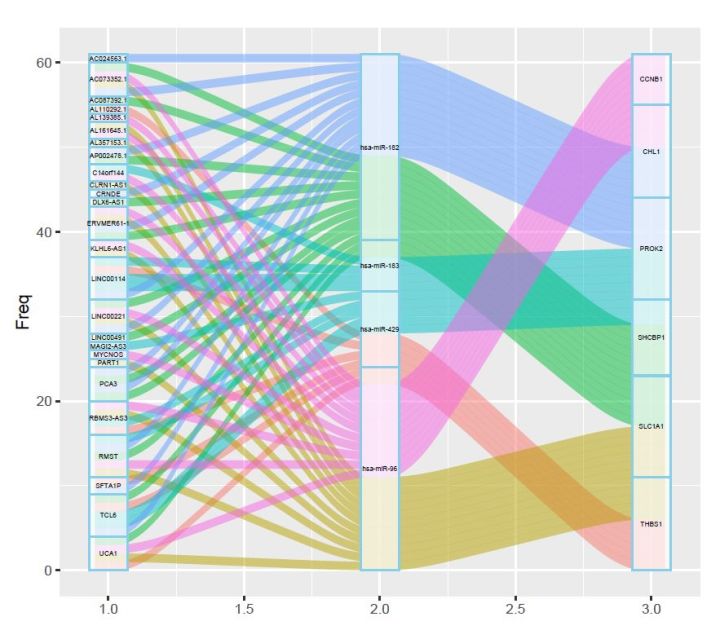
# 绘制stratum,添加文字标签并隐藏图例;
p2<-p1+geom_stratum(fill="white",color="skyblue",alpha=.7,width =1/7)+
geom_text(stat ="stratum",size =1.5,color="black",label.strata =T)+
guides(fill =FALSE)
p2
#修改横轴的标签;
p3<-p2+
scale_x_discrete(limits = c("lncRNA","miRNA","mRNA"),expand = c(0, 0))+
xlab("") + ylab("")
#自定义主题;
mytheme1<-theme_bw() +
theme(panel.grid =element_blank()) +
theme(panel.border = element_blank()) +
theme(axis.line = element_blank(),axis.ticks =element_blank(),axis.text.y =element_blank())
p4<-p3+mytheme1
p4
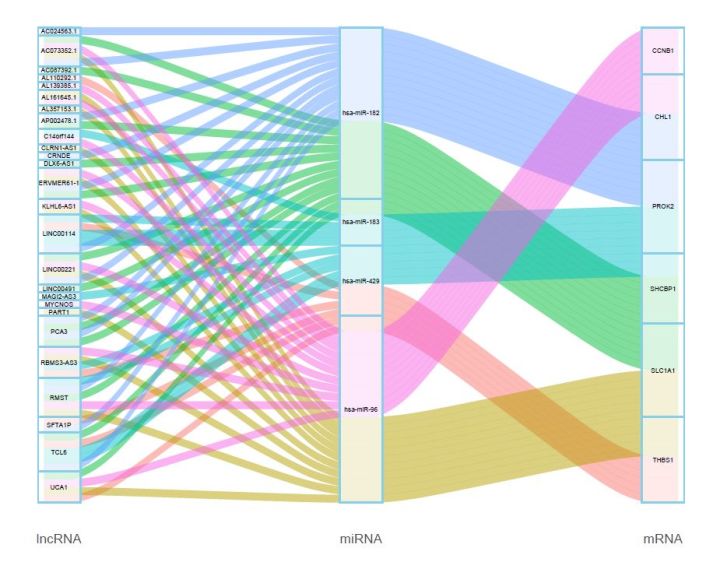
“长数据”作图
使用宽数据画图比较直观,容易理解,但如果想给不同的stratum添加不同的颜色就不是很方便。其实,ggalluvial也支持长数据,毕竟是基于ggplot2的。
#检查数据是否符合要求;
head(df)
is_alluvia_form(df,weight ="Freq")
# 转成长数据格式;
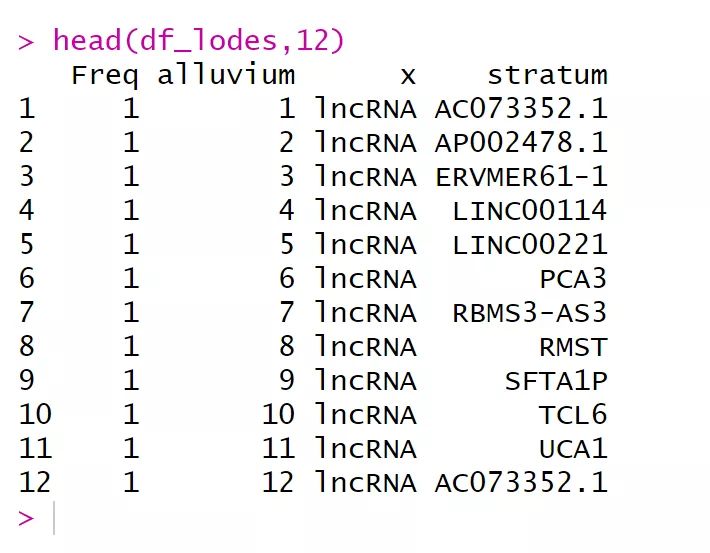
df_lodes <- to_lodes_form(df,key ="x", value = "stratum", id = "alluvium",axes =1:3)
#检查转换后的数据是否符合作图要求;
head(df_lodes,12)
is_lodes_form(df_lodes,key = "x",value = "stratum",id = "alluvium",weight ="Freq")
#自己生成渐变色;
mycol3=colorRampPalette(c("#00abef","#64b036","#ffe743","#64b036","#00abef"))(36)
对于绘图方法,除了数据映射方式,其他参数的用法与上文相似,这里就不做代码分解演示啦。
p5<-ggplot(df_lodes,aes(x = x, stratum =stratum, alluvium = alluvium,
fill = stratum, label = stratum)) +
scale_x_discrete(expand = c(0, 0)) +
geom_flow(width = 0.2, knot.pos = 0.1) +
geom_stratum(alpha = .9,color="grey20",width = 1/7) +
geom_text(stat = "stratum", size =1.5,color="black") +
scale_fill_manual(values = mycol3) +
xlab("") + ylab("") +
theme_bw() +
theme(panel.grid =element_blank()) +
theme(panel.border = element_blank()) +
theme(axis.line = element_blank(),axis.ticks =element_blank(),axis.text.y =element_blank())+
guides(fill = FALSE)
p5
绘图的结果如下:
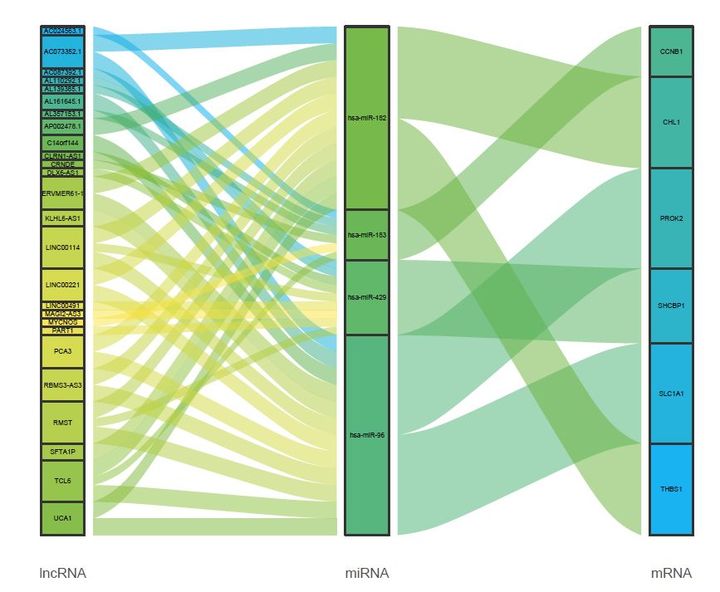
如果用ggplot2的默认配色,其实也挺好看的,如下:
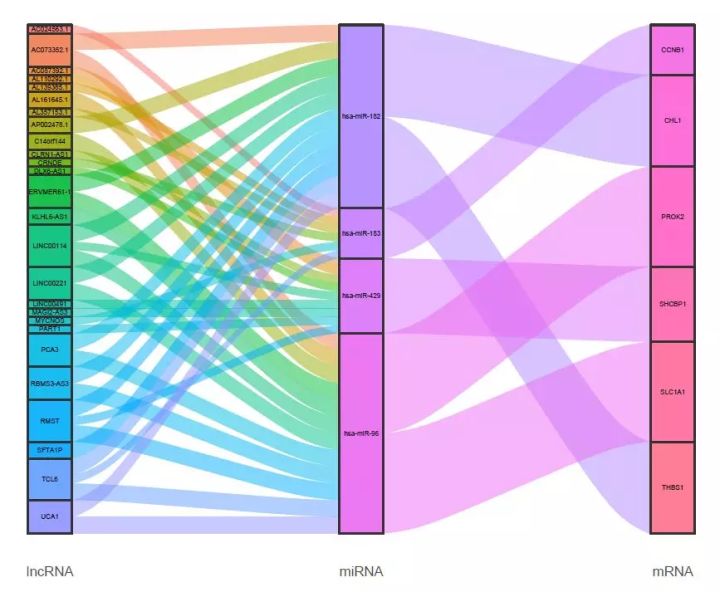
#使用文章中图表的颜色集;
mycol <-rep(c("#223D6C","#D20A13","#FFD121","#088247","#11AA4D","#58CDD9","#7A142C","#5D90BA","#029149","#431A3D","#91612D","#6E568C","#E0367A","#D8D155","#64495D","#7CC767","#223D6C","#D20A13","#FFD121","#088247","#11AA4D","#58CDD9","#7A142C","#5D90BA","#029149","#431A3D","#91612D","#6E568C","#E0367A","#D8D155","#64495D","#7CC767","#223D6C","#D20A13","#FFD121","#088247","#11AA4D","#58CDD9","#7A142C","#5D90BA","#029149","#431A3D","#91612D","#223D6C","#D20A13","#FFD121","#088247","#11AA4D","#58CDD9","#7A142C","#5D90BA","#029149","#431A3D","#91612D","#6E568C","#E0367A","#D8D155","#64495D","#7CC767","#223D6C","#D20A13","#FFD121","#088247","#11AA4D","#58CDD9","#7A142C","#5D90BA","#029149","#431A3D","#91612D","#6E568C","#E0367A","#D8D155","#64495D","#7CC767"),3)
如果想使用文章中的颜色,可将mycol变量赋值给scale_fill_manual(values = mycol);同时不想让stratum周围显示白色的“空白”,可将geom_flow()的width设为小于0.2的值, 得到效果如下:
您好,想请教一下老师,什么是“宽数据”和“窄数据”呢?以及,我该如何构建绘制sankey图的“宽数据”或者“窄数据”,这些数据里面需要包含哪些信息呢?
祝科研顺
你好,我有一篇关于宽窄数据的转换,你可以看一下ggplot2画图之前的准备—-数据宽长变换