- 使用ggplot2绘制LD50图
library(ggplot2)
### this is the data ###
# data from four experiments
conc <- c(5.00E-07, 1.00E-06, 1.00E-05,
5.00E-07, 1.00E-06, 5.00E-06, 1.00E-05, 2.00E-05,
5.00E-07, 1.00E-06, 2.50E-06, 5.00E-06, 1.00E-05,
5.00E-07, 1.00E-06, 2.50E-06, 5.00E-06, 1.00E-05)
dead.cells <- c(34.6, 47.7, 81.7,
37.6, 55.7, 89.1, 84.3, 85.2,
34.4, 46.1, 76.2, 84.3, 84.1,
24.5, 26.1, 60.6, 82.7, 87)
# transform the data to make it postive and put into a data frame for fitting
data <- as.data.frame(conc) # create the data frame
data$dead.cells <- dead.cells
data$nM <- data$conc * 1000000000
data$log.nM <- log10(data$nM)
### fit the data ###
# make sure logconc remains positive, otherwise multiply to keep positive values
# (such as in this example where the iconc is multiplied by 1000
fit <- nls(dead.cells ~ bot+(top-bot)/(1+(log.nM/LD50)^slope),
data = data,
start=list(bot=20, top=95, LD50=3, slope=-12))
m <- coef(fit)
val <- format((10^m[3]),dig=0)
### ggplot the results ###
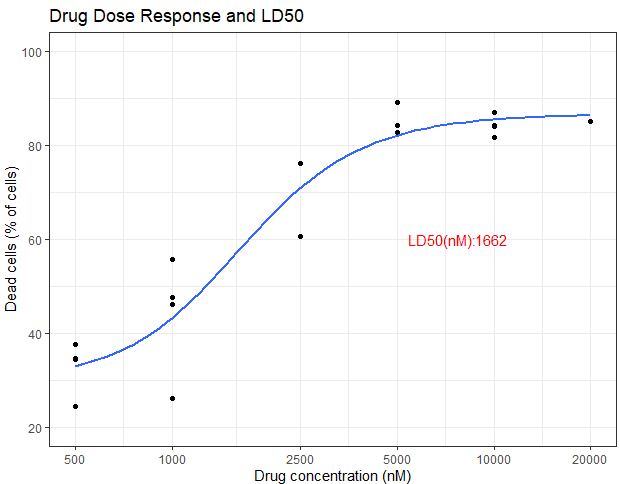
p <- ggplot(data=data, # specify the data frame with data
aes(x=nM, y=dead.cells)) + # specify x and y
geom_point() + # make a scatter plot
scale_x_log10(breaks = c(500, 1000, 2500, 5000, 10000, 20000))+
xlab("Drug concentration (nM)") + # label x-axis
ylab("Dead cells (% of cells)") + # label y-axis
ggtitle("Drug Dose Response and LD50") + # add a title
theme_bw() + # a simple theme
expand_limits(y=c(20,100)) # customise the y-axis
# Add the line to graph using methods.args (New: Jan 2016)
p <- p + geom_smooth(method = "nls",
method.args = list(formula = y ~ bot+(top-bot)/(1+( x / LD50)^slope),
start=list(bot=20, top=95, LD50=3, slope=-12)),
se = FALSE)
# Add the text with the LD50 to the graph.
p <- p+ annotate(geom="text", x=7000, y= 60, label="LD50(nM): ", color="red") +
annotate(geom="text", x=9800, y= 60, label=val, color="red")
p # show the plot
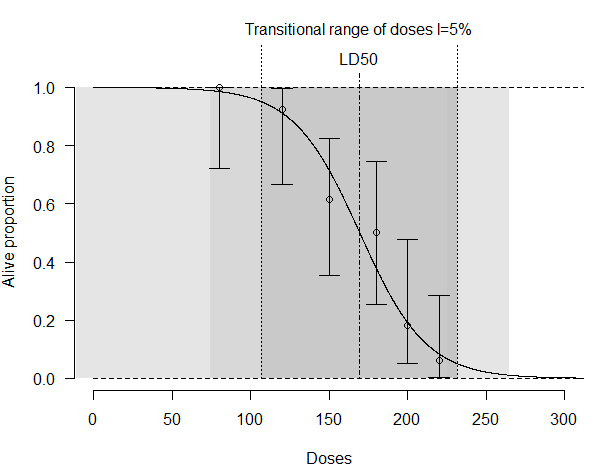
2. LD50 function from HelpersMG package
library("HelpersMG")
data <- data.frame(Doses=c(80, 100, 120, 150, 180, 200),
Alive=c(10, 12, 8, 6, 2, 1),
Dead=c(0, 1, 5, 6, 9, 15))
LD50_logistic <- LD50(data, equation="logistic")
predict(LD50_logistic, doses=c(140, 170))
plot(LD50_logistic, xlim=c(0, 300), at=seq(from=0, to=300, by=50))
LD50_probit <- LD50(data, equation="probit")
predict(LD50_probit, doses=c(140, 170))
plot(LD50_probit)
LD50_logit <- LD50(data, equation="logit")
predict(LD50_logit, doses=c(140, 170))
plot(LD50_logit)
LD50_hill <- LD50(data, equation="hill")
predict(LD50_hill, doses=c(140, 170))
plot(LD50_hill)
LD50_Richards <- LD50(data, equation="Richards")
predict(LD50_Richards, doses=c(140, 170))
plot(LD50_Richards)
LD50_Hulin <- LD50(data, equation="Hulin")
predict(LD50_Hulin, doses=c(140, 170))
plot(LD50_Hulin)
LD50_DoubleRichards <- LD50(data, equation="Double-Richards")
predict(LD50_DoubleRichards, doses=c(140, 170))
plot(LD50_DoubleRichards)
LD50_flexit <- LD50(data, equation="flexit")
predict(LD50_flexit, doses=c(140, 170))
plot(LD50_flexit)