原理
因为每个基因的各区都不一样长,所以需要将reads对区域长度进行归一化。针对参考基因组的3’UTR,CDS以及5’UTR分别设置独立的coordinate进行归一化处理,然后再将bed或者bam文件map回去,最后绘制plotprofile。经过阅读paper和多方比较,可以通过R包Guitar(适用于MeRIP-Seq,BS-Seq,一般的RNA-Seq and etc.)快速实现上述要求。
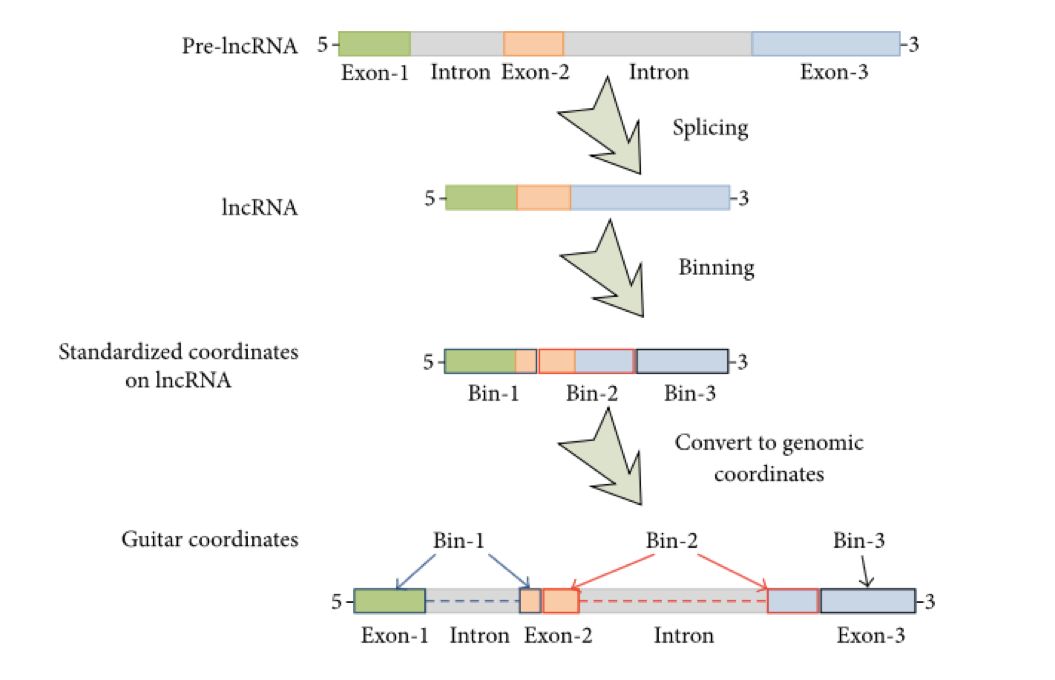
原理图
过程:
1.R包的安装
这个R包是发布于bioconductor网站(一个强大的开源生信软件资源网站),安装过程不同于一般R包的安装.
运行R:
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("Guitar")安装后查看包的安装情况以及documentation
browseVignettes("Guitar")2. 下载基因组文件
library(Guitar)生成包含基因注释信息的TxDb object,可以使用GenomicFeatures包从UCSC获取基因注释文件,示例中选择的是mm10
#安装GenomicFeatures包
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("GenomicFeatures")
#提示需要安装RMariaDB包,这个包需要系统上安装mysql环境
install.packages("RMariaDB")
library(RMariaDB)
library(GenomicFeatures)
txdb <- makeTxDbFromUCSC(genome="mm10")Download the knownGene table ... OK
Download the knownToLocusLink table ... OK
Extract the 'transcripts' data frame ... OK
Extract the 'splicings' data frame ... OK
Download and preprocess the 'chrominfo' data frame ... OK
Prepare the 'metadata' data frame ... OK
Make the TxDb object ... OK3.建立coords,这里的coords被命名为Guitar coordinates
gc_txdb <- makeGuitarCoordsFromTxDb(txdb, noBins = 20)[1] "total 63759 transcripts extracted ..."
[1] "total 49758 transcripts left after ambiguity filter ..."
[1] "total 19295 mRNAs left after component length filter ..."
[1] "total 7695 ncRNAs left after ncRNA length filter ..."
[1] "Building Guitar Coordinates. It may take a few minutes ..."
[1] "Guitar Coordinates Built ..."4.载入待处理的bed和bam文件
bedrange <- import.bed(bed)# bed imported as GRange
bamalignment <- readGAlignments(bam) #bam imported as GRangelist
feature_mm10 <- list(bedrange,bamalignment)
names(feature_mm10) <- c("bedrange","bamalignment")5.基于前两步生成的文件,生成plot
GuitarPlot(gfeatures=feature_mm10,GuitarCoordsFromTxDb = gc_txdb,saveToPDFprefix = "example")[1] "Using provided Guitar Coordinates"
[1] "resolving ambiguious features ..."
[1] "Figures saved into example_Guitar.pdf ..."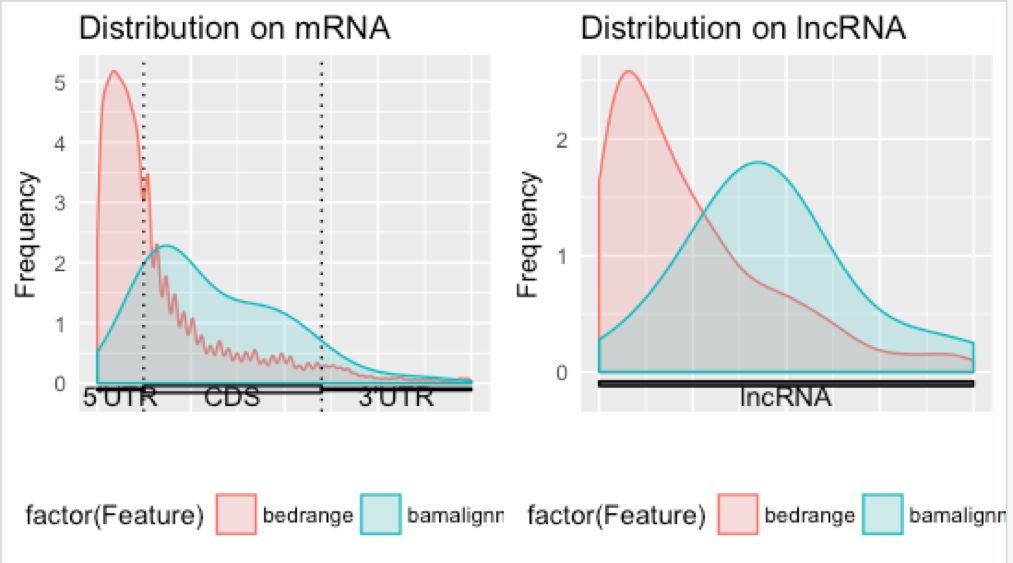
结果图
- 附:
可以通过Guitar包实现的其他例子:
参考文献/网址:
- Guitar: An R/Bioconductor Package for Gene Annotation Guided
- Transcriptomic Analysis of RNA-Related Genomic Features
- https://www.hindawi.com/journals/bmri/2016/8367534/
- http://bioconductor.org/packages/release/bioc/html/Guitar.html
您好,请问为何使用makeGuitarCoordsFromTxDb函数会出现:Error in makeGuitarCoordsFromTxDb(txdb, noBins = 20): could not find function “makeGuitarCoordsFromTxDb” 这个错误呢?谢谢!
你好,程序包有更新,函数简化了,具体参考官方文档,我也整理过来了,请参考:UTR区域的分布plotprofile——R包Guitar