涉及拼图时,有时几个图图例并不完全一样,这时候如何处理,直接上代码(以pancancer生存分析数据为例),此处用到lomon和gridExtra包
library(ggplot2)
setwd("G:/test")
unicox1 <- read.csv("OS_pancan_unicox.csv")
unicox1 <- unicox1[order(unicox1$HR,decreasing = F),]
unicox1$cancer <- factor(unicox1$cancer, levels = unicox1$cancer)
unicox2 <- read.csv("PFI_pancan_unicox.csv")
unicox2$cancer <- factor(unicox1$cancer, levels = unicox1$cancer)
unicox3 <- read.csv("DSS_pancan_unicox.csv")
unicox3$cancer <- factor(unicox1$cancer, levels = unicox1$cancer)
unicox4 <- read.csv("DFI_pancan_unicox.csv")
unicox4$cancer <- factor(unicox1$cancer, levels = unicox1$cancer)
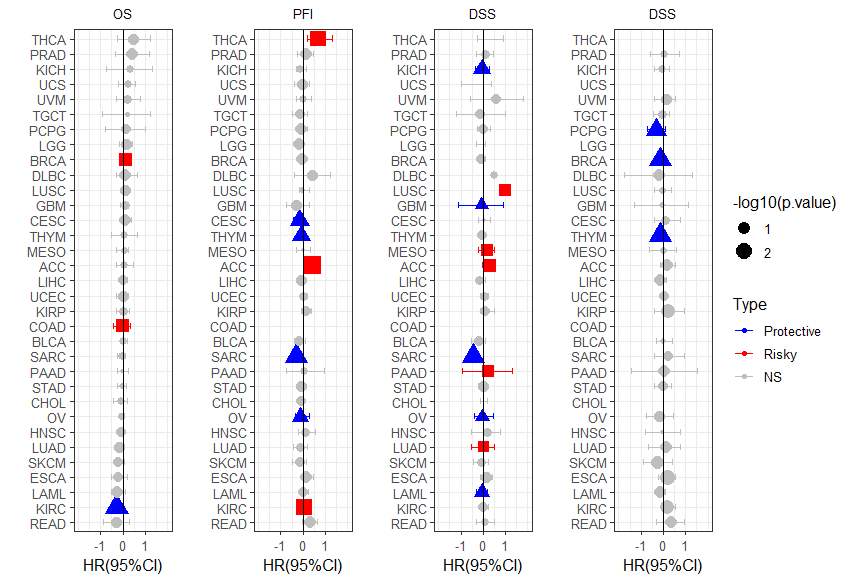
p1 <- ggplot(unicox1, aes(HR, cancer, col=Type,shape=Type))+
geom_point(aes(size=-log10(p.value)))+
geom_errorbarh(aes(xmax =upper_95, xmin = lower_95), height = 0.4)+
scale_x_continuous(limits= c(-2, 2), breaks= seq(-1, 1, 1))+
geom_vline(aes(xintercept = 0))+
xlab('HR(95%CI)') + ylab(' ')+
theme_bw(base_size = 12)+
ggtitle("OS")+
theme(plot.title = element_text(hjust = 0.5, size=10))+
scale_color_manual(values = c("gray", "blue", "red"))+theme(legend.position='none')
p2 <- ggplot(unicox2, aes(HR, cancer, col=Type,shape=Type))+
geom_point(aes(size=-log10(p.value)))+
geom_errorbarh(aes(xmax =upper_95, xmin = lower_95), height = 0.4)+
scale_x_continuous(limits= c(-2, 2), breaks= seq(-1, 1, 1))+
geom_vline(aes(xintercept = 0))+
xlab('HR(95%CI)') + ylab(' ')+
theme_bw(base_size = 12)+
ggtitle("PFI")+
theme(plot.title = element_text(hjust = 0.5, size=10))+guides(shape=FALSE) +
scale_color_manual(values = c( "blue", "red","gray"),
breaks = c("Protective","Risky","NS"),
labels = c("Protective","Risky","NS"))+theme(legend.position='none')
p3 <- ggplot(unicox3, aes(HR, cancer, col=Type,shape=Type))+
geom_point(aes(size=-log10(p.value)))+
geom_errorbarh(aes(xmax =upper_95, xmin = lower_95), height = 0.4)+
scale_x_continuous(limits= c(-2, 2), breaks= seq(-1, 1, 1))+
geom_vline(aes(xintercept = 0))+
xlab('HR(95%CI)') + ylab(' ')+
theme_bw(base_size = 12)+
ggtitle("DSS")+
theme(plot.title = element_text(hjust = 0.5, size=10))+
scale_color_manual(values = c("gray", "blue", "red"))+theme(legend.position='none')
p4 <- ggplot(unicox4, aes(HR, cancer, col=Type,shape=Type))+
geom_point(aes(size=-log10(p.value)))+
geom_errorbarh(aes(xmax =upper_95, xmin = lower_95), height = 0.4)+
scale_x_continuous(limits= c(-2, 2), breaks= seq(-1, 1, 1))+
geom_vline(aes(xintercept = 0))+
xlab('HR(95%CI)') + ylab(' ')+
theme_bw(base_size = 12)+
ggtitle("DSS")+
theme(plot.title = element_text(hjust = 0.5, size=10))+
scale_color_manual(values = c("gray", "blue", "red"))+theme(legend.position='none')
library(gridExtra)
library(lemon)
legend <- g_legend(p2)
grid.arrange(p1, p2,p3,p4,
ncol = 4, nrow = 1,right=legend)
进哥您好!我想请问一下可不可以分享一期单基因泛癌的COX分析教程呀?